Brian Williamson and Subdoh Nimkar
Applied Biosystems, Framingham, MA, USA
Introduction
Mass spectrometry-based tagging methods enable the relative expression levels of large sets of proteins to be measured and quantitated with a high degree of automation. One of these technologies uses isobaric tagging for the simultaneous identification and quantification of proteins from multiple samples in a single multiplex analysis.1 The isobaric peptide tagging makes it possible to pool protein samples without increasing the complexity of the mass spectrometry (MS) analysis, or losing important post-translational modification (PTM) information. A new set of isobaric tagging reagents has recently become available that, for the first time, allows up to eight different protein samples to be multiplexed (8plex), doubling the number of states that can be compared.2 This provides greater flexibility in the design of experiments, such as by allowing more experimental samples to be compared within a single run or permitting inclusion of greater numbers of duplicates within the same sample for increased statistical support. The 8plex approach also has the potential to increase sample throughput while reducing the costs of time and labour.
The new 8plex reagents, iTRAQ reagents (Applied Biosystems), are amine-specific, stable isotope reagents that use the same chemistry as the 4plex reagents.1 Multiple peptides per protein can be tagged, increasing confidence in identification and quantitation from MS/MS spectra. Identical peptides labelled with the different reagents exhibit the same parent ion; MS/MS fragmentation of the parent ion generates unique signature ions that distinguish the individual samples, and the relative abundance among the samples can then be determined (Figure 1). The 8plex reporter ions (isobaric tags) are 113, 114, 115, 116, 117, 118, 119 and 121.2
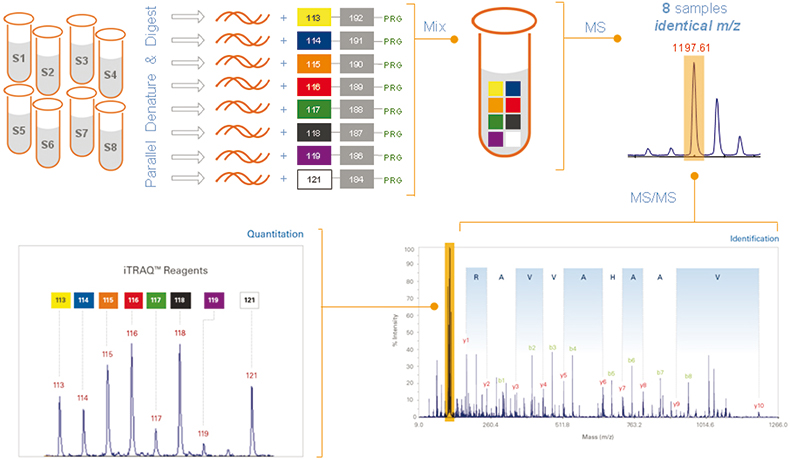
Figure 1. Workflow:3 once labelled with the isobaric iTRAQ reagents–8plex, the eight individual digested protein samples are pooled for further processing and MS analysis. Each peptide is detected by MS and then identified by MS/MS analysis. In addition, each isobaric tag produces a unique reporter ion in the MS/MS spectrum that identifies which samples the peptide originated from and its relative abundance.
The effectiveness of these new reagents in quantitating eight states simultaneously has been determined against a set of known peptides and proteins,3 and is outlined in this article. The reagents were evaluated for label efficiency, fragmentation efficiency and precision and accuracy of quantitation.
Labelling efficiency
A time course study was carried out on a three-peptide mix. Three peptides were selected that contained single, double and triple labelling sites [*LNENIR, *ETLDPSAPK* and *LSLGGLLQPEK*PVVLK*; (* denotes potential labelling site)]. The three-peptide mix was first labelled with the 8plex reagent 113 reporter ion for 30 minutes, then the 115 label was added and reacted for 30 minutes; this was repeated with three additional reagents. MS/MS of the labelled peptide *LSLGGLLQPEK*PVVLK* showed strong presence of the 113 reporter ion; the 115, 117, 119 and 121 reporter ions were absent, suggesting complete labelling after treatment with the 113 reagent (see Figure 2).
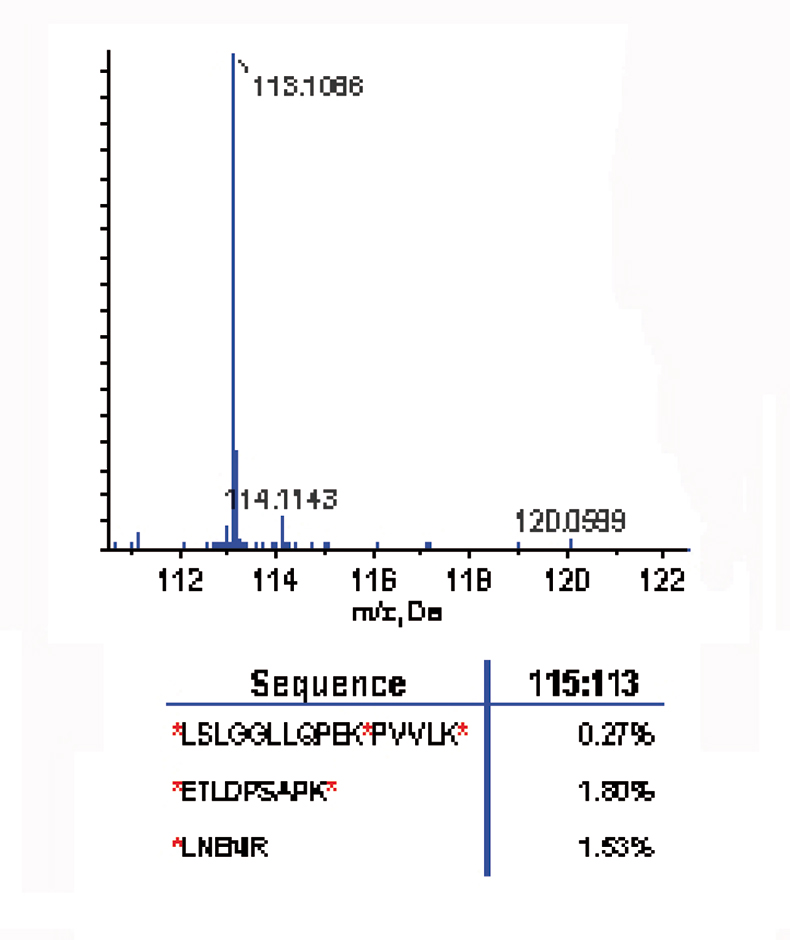
Figure 2. Labelling efficiency: the reporter ion region (top) from the MS/MS labelled peptide (*LSLGGLLQPEK*PVVLK*) shows the strong presence of the 113 reporter ion and the absence of the 115, 117, 119 and 121 reporter ions, suggesting complete labelling after treatment with the 113 reagent.3 The relative ratios of the reporter ion for all three peptides (bottom) demonstrate that the reaction was 98% complete after 30 minutes.
Accuracy and precision of relative quantitation
Carbonic anhydrase and ovotransferrin were spiked at differing concentrations into eight vials of a six-protein mix. The samples were digested, labelled with the 8plex reagents in triplicate and analysed on a QSTAR Elite LC/MS/MS system (Applied Biosystems). The observed ratios from all eight proteins were close to the expected values.3 The coefficient of variance (CV) was calculated for the observed serotransferrin peptides and the average CV for all eight reagents was 14%.
Signal amplification
A six-protein mix was labelled with all eight 8plex reagents and two pools were created. Ovotransferrin labelled with only the 113 8plex reagent was added to the first pool, and ovotransferrin labelled with all eight of the reagents was spiked into the second pool. This is an eight-fold increase in the overall concentration of ovotransferrin, but the concentration of ovotransferrin labelled with the 113 reagent remained constant. LC/MS/MS analysis of the two pools showed a clear amplification of the MS/MS fragment ions and an increase in ovotransferrin protein coverage (from 29% to 63%).3
Conclusions
The ways in which protein expression profiles from complex protein mixtures can be studied are rapidly expanding within the area of proteomics.2,4 The multiplex 8plex peptide-labelling reagents are a powerful method for accurate protein quantitation. The unique isobaric nature of the reagents allows up to eight samples to be processed simultaneously, with over 98% labelling efficiency. These reagents are an effective and reproducible method for quantifying differentially expressed proteins.
References
- P.L. Ross, Y.N. Huang, J.L. Marchese, B. Williamson, K. Parker, S. Hattan, N. Khainovski, S. Pillai, S. Dey, S. Daniels, S. Purkayastha, P. Juhasz, S. Martin, M. Bartlet-Jones, F. He, A. Jacobson and D.J. Pappin, “Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents”, Mol. Cell. Proteomics 3, 1154–1169 (2004). https://doi.org/10.1074/mcp.M400129-MCP200
- L. Choe, M. D’Ascenzo, N.R. Relkin, D. Pappin, P. Ross, B. Williamson, S. Guertin, P. Pribil and K.H. Lee, “8-Plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobin treatment for Alzheimer’s disease”, Proteomics 7, 3651–3660 (2007). https://doi.org/10.1002/pmic.200700316
- Multiplex protein quantitation using iTRAQ® Reagents–8plex. Technical note, available from Applied Biosystems (www.appliedbiosystems.com).
- M. D’Ascenzo, L. Choe and K.H. Lee, “iTRAQPak: an R based analysis and visualization package for 8-plex isobaric protein expression data”, Brief. Funct. Genomic Proteomic., published online 13 February 2008.